One-step RT-PCR reactions consistently use a gene-specific primer for first-strand cDNA synthesis, while two-step RT-PCR reactions permit for additional priming choices.
“When the mRNA includes a poly-A 3′ tail, then a oligo-dT primer may be used to prime each of mRNAs simultaneously.
SSPs give the best specificity and also have been proven to be the most persistent of the primer selections for reverse transcription. But they don’t offer you the flexibility of oligo(dT) and random primers, meaning a fresh cDNA synthesis reaction has to be done for each gene to be analyzed. This creates primers for processing restricting cell or tissue samples.
Oligo(dT) primers aren’t recommended as the sole real primer for cDNA synthesis of 18S rRNA can be used for normalization at a real-time PCR experimentation as the oligo(dt) primer won’t anneal.
To create cDNA in the subset of mRNA, afterward a sequence-specific primer can be utilized that will only bind to a mRNA sequence.
Multiple kinds of oligo(dT) primers can be found. Finally, anchored oligo(dT) primers are made to steer clear of polyA slippage by making sure they anneal in the three ´UTR/polyA junction. Deciding on the most effective oligo(dT) primer can rely in part on the warmth of the reverse transcription. Thermostable RTs like SuperScript® III Reverse Transcriptase may function better with primers, which stay annealed in comparison for their counterparts.
Sequence Particular Primers
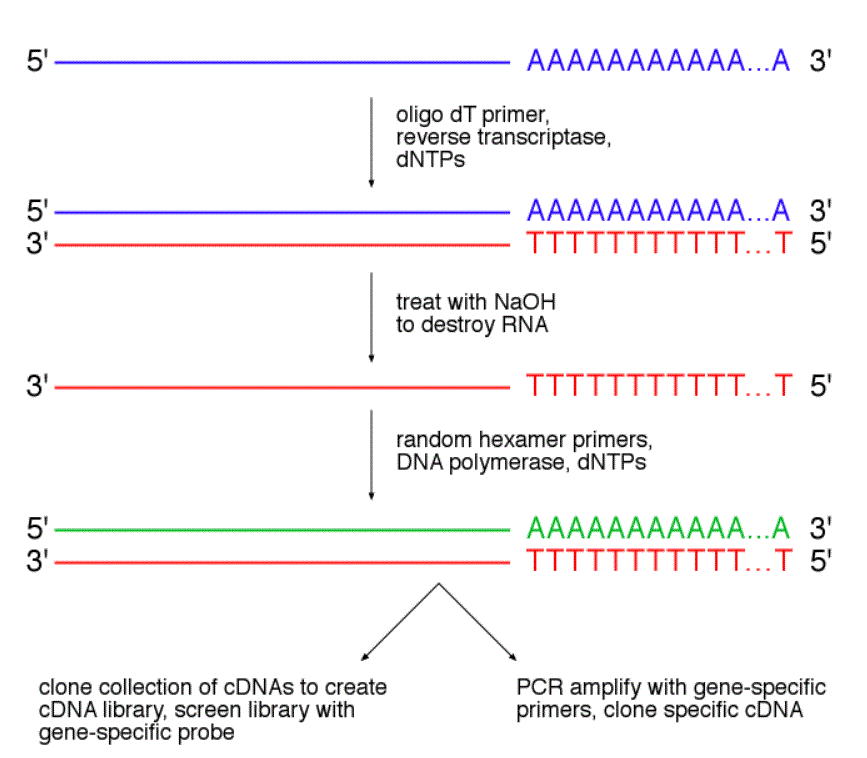
http://www.bio.davidson.edu/courses/genomics/method/cdnaproduction.html
Implementing a mix of random and oligo(dT) primers can at times raise information quality by combining the advantages of both if utilized in precisely the exact same first-strand cDNA synthesis reaction. Random primers are utilized in reactions.
Oligo(dT) primers are a favourite choice for two-step cDNA synthesis reactions due to their specificity for both mRNA and since they permit several distinct targets to be analyzed by precisely the exact same cDNA pool. Since they initiate transcription structure that is hard can lead to cDNA synthesis.
In case you wanted to create parts of cDNA which were scattered all around the mRNA, then you may use an arbitrary primer cocktail which could create cDNA from many mRNAs however, the cDNAs wouldn’t be full length. The advantages to priming would be the creation of cDNA fragments that are shorter and raising the likelihood that 5′ ends of the mRNA will be converted into cDNA. Since reverse transcriptase does not usually achieve the 5′ ends of extended mRNAs, arbitrary primers may be beneficial”
Since they anneal through the target molecule They’re also excellent for RNA, such as RNA.
