Understanding cell death has a huge impact on the treatment of diseases. Apoptosis is a genetically determined suicide program that removes unnecessary or potentially harmful cells. Apoptosis is an important process in embryonic development, cell aging, the immune response and the response to poisoning. Deviations in this program can lead to neurodegenerative or autoimmune diseases. Blocking apoptosis is also one of the “hallmarks of cancer” postulated by Hanahan and Weinberg. 1
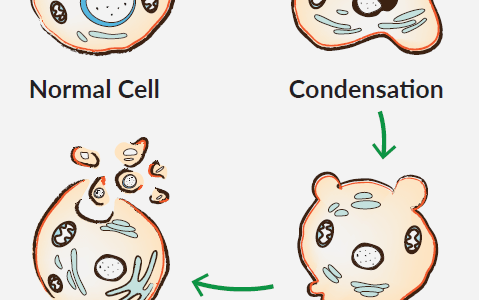
Apoptosis leads to a clear morphological change in the cell due to a non-inflammatory cascade of molecular events. These include the reduction of cell volume, the fragmentation of the cell nucleus, the condensation of chromatin, the degradation of DNA, the formation of bubbles on the cell membrane and the subsequent formation of apoptosis bodies. This sequence not only leads to the destruction of unwanted cells, but also prepares the cell debris for removal by phagocytes. In view of the variety of apoptotic stimuli and their effects on different signaling pathways, it is important to understand what exactly led to cell death in an experimental setup.
Mitochondrial membrane potential
Mitochondria are involved in the apoptotic process in several places. During the life of a cell, mitochondria use oxidizable substrates to create a proton gradient along the inner mitochondrial membrane. During apoptosis, this membrane potential decreases in connection with the opening of the mitochondrial permeability pores and the release of apoptogenic factors such as cytochrome C. In some apoptotic models, the loss of membrane potential is considered an early event in the apoptotic process. Other researchers suspect that the loss of membrane potential is a result of the apoptotic signaling cascade. 2-4 An easy way to assess the membrane potential of mitochondria in a cell population is to use positively charged dyes such as JC-1 and TMRE, which are found in the electronegative Collect inside active mitochondria.
JC-1 / TMRE
JC-1 changes from red fluorescence in healthy mitochondria to green fluorescence when, as in apoptosis, the membrane potential is lost. TMRE accumulates in polarized mitochondria and is particularly suitable for observing the membrane potential of living cells. Depolarized mitochondria with reduced membrane potential are unable to accumulate TMRE.
Caspase activation
The key figures in the apoptotic signaling pathway are cysteine-dependent ASpartate-specific ProteASEN (caspases), the efficient activation of which determines the fate of the cell. Caspases are activated by several signal paths and form reinforcing loops. The external signaling pathway binds extracellular death ligands such as FasL or TNF-α to transmembrane death receptors, which in turn recruit initiator caspase-8. In the inner signal pathway, the cytochrome C released by mitochondria can trigger the formation of the caspase-activating complex (also called apoptosome). The latter in turn recruits and activates the initiator caspase-9. The initiator caspases then cleave and activate the effector caspases 3 and 7 and lead to the cleavage of specific substrates, which leads directly to the morphological changes that traditionally define cellular apoptosis.5 This is a measurement of the caspase activity important indicator of the ongoing apoptotic process.
Caspase 3/7
The protease activity of caspases 3 and 7 can be detected using the fluorogenic substrate N-Ac-DEVD-N’-MC-R110, which produces a fluorescent product on cleavage.
Core fragmentation, chromatin condensation and DNA degradation
Caspases also break down internal cell structures and help with their efficient disposal. One of the most remarkable events in this process is the condensation of the cell nucleus and its breakdown into smaller fragments. The core fragmentation results from the dissolution of the nuclear lamina after proteolysis by caspases and the collapse of the nuclear envelope. Another characteristic of apoptosis is the condensation of chromatin, accompanied by hydrolysis of the core DNA. Hoechst 33342, a cell-permeable, fluorescent DNA dye, is often used to microscopically analyze chromatin condensation. The Golgi, the endoplasmic reticulum and the mitochondrial network also disintegrate during apoptosis. During the breakdown, the fragments are distributed in bubbles along the plasma membrane.
DNA dyes
Hoechst dyes are cell-compatible and bind nucleic acids in living and fixed cells. Its blue, and therefore little overlapping, emission spectrum makes it recommended for researchers who plan to carry out several fluorescent stains on a sample. An alternative cell-compatible DNA dye is DRAQ5 ™, whose deep red excitation spectrum can be combined with fluorophores that emit blue and orange light. Non-membrane-compatible dyes such as propidium iodide, DAPI, DRAQ7 ™ or RedDot ™ 2 are ideal for the specific staining of the nuclei of dead cells where the integrity of the plasma membrane is impaired.
Blistering
Caspases also cleave many major components of the cytoskeleton, thereby rounding and retracting the cells, which is typical of early stages of apoptosis. Another result of the weakening of the cytoskeleton is the formation of membrane vesicles. When the cytoplasm presses against weakened areas of the plasma membrane, these show themselves as bulges that can be visualized microscopically. These vesicles are believed to be a result of myosin-dependent contraction of cortical actin bundles that press the cytoplasm against the cell cortex. Blistering on the plasma membrane is an important step on the path of the apoptotic cell towards the smaller apoptotic bodies.
Elimination of apoptotic cells
A critical component of the apoptotic process is the complete lack of an inflammatory response to dying cells. The disintegration of the apoptotic cells into apoptotic bodies prevents the release of the Damage-Associated Molecular Patterns (DAMPs) into the extracellular space and thereby facilitates the disposal by phagocytes. In early apoptosis, apoptotic cells secrete a “find me” signal and later a “eat me” signal to recruit phagocytes. This targeted interaction between apoptotic cells and phagocytes ensures that dying cells are eliminated by the non-inflammatory signaling pathway. “Find me” signals that recruit phagocytes include lysophosphatidylcholine, sphingosine-1-phosphate, fractal kinine and nucleotides such as ATP and UTP. These nucleotides are released through a caspase-mediated channel opening of the pannexin-1 (PANX1) channels.6 The caspase-dependent PANX1 channel opening also allows a small group of fluorescent monomeric cyanine dyes such as TO-PRO®-3 to flow in. In this way, the pannexin channels can be used to identify cells that send “find me” signals. 7
As soon as phagocytes approach an apoptotic cell, they recognize phosphatidylserine and phosphatidylethanolamine residues that come from inside the phospholipid double membrane and are now exposed on the cell surface. This rearrangement, which is typical of apoptotic cells, distinguishes it from its viable counterparts and provides the phagocytes with a “eat me” signal. The phospholipid-binding protein Annexin V adheres to phospholipid residues that are exposed during apoptosis. It is used to detect phosphatidylserine on the outer membrane of apoptotic cells and to determine the “eat me” phase of apoptosis.8,9
TO-PRO®-3
TO-PRO®-3 is a deep red fluorescent dye that enables the detection of an early event of apoptosis. The dye uses the caspase-dependent activated pannexin channels to penetrate the cells. This makes it possible to identify apoptotic cells that cannot be detected with the help of traditional markers such as Annexin V due to the process that has just started.
Annexin V
Annexin V can be conjugated with various fluorochromes (e.g., FITC, PE, APC) to stain phosphatidylserine on the outer plasma membrane. The detection with Annexin V can be combined with other markers, such as those for membrane integrity (for example, RedDot ™ 2, propidium iodide, DAPI), in order to distinguish apoptotic from necrotic cells.
Multiplex is the key
The above-mentioned morphological features of the apoptotic signaling pathway can help determine the type of cell death in a model system. However, a single measurement can lead to misinterpretations for certain experimental questions (e.g. the distinction between necrosis and apoptosis). In contrast to single measurements, multiplex assays provide a more complete picture of the apoptotic process. For example, the combined measurement with TO-PRO®-3, Annexin V, TMRE and DAPI allows a simultaneous statement on “Find me” or “Eat me” signals, the mitochondrial membrane potential and the core fragmentation. Multiplex assays thus provide a complex picture of the apoptotic processes and how they affect cells. In addition, markers for early apoptosis (eg TO-PRO®-3) can be used to identify cells that were considered viable due to the limitations of traditional apoptosis markers (eg Annexin V). Muliplex methods thus, by using different markers for the different phases of apoptosis, enable a quantitative classification of the entire cell population into the stages: viable, early apoptosis, late apoptosis, apoptosis bodies and non-cellular debris.
The selection of cell-based assays from Cayman Chemical provide you with flexible and efficient tools for determining the different stages of cell death in your model system. Biomol’s technical support and Cayman’s product developers will be happy to help you choose the cell-based assay that suits you, so that you are able to get the most accurate answers to your research questions.